When scientists discovered the gene that causes cystic fibrosis in 1989, they were optimistic that a cure was on the horizon. As the years passed by, hundreds of mutations were identified in the gene, but new treatments were slow to emerge and a cure has yet to materialize. One major reason for the delay is that scientists have had trouble figuring out precisely where the gene is active. Now they know.
A multidisciplinary team of researchers at the Novartis Institutes for BioMedical Research (NIBR) and Harvard Medical School (HMS) worked to catalog all the different cells in the airway, and the paths they take to become those cells. In the process, they discovered a completely new type of cell, which they named a pulmonary ionocyte. When the cystic fibrosis gene from 1989 – dubbed CFTR – is active, it is usually in the pulmonary ionocyte, which makes up just 1-2% of the airway. The team reported this discovery online in Nature on August 1. A similar finding was reported by scientists at the Broad Institute and Massachusetts General Hospital, in the same journal on the same day.
“We can use this information to be a bit more clever when we devise therapeutic approaches to cystic fibrosis,” says Aron Jaffe, co-corresponding author and a co-leader of respiratory disease research at NIBR.
“As people work toward cures, knowing you are looking at 1% of the cell population seems essential for any type of troubleshooting to improve a therapy or develop new therapies,” adds Allon Klein, co-corresponding author and assistant professor of systems biology at HMS.
Noting that many of the CFTR mutations only stop part of the gene’s function, Jaffe suggests that for those mutations, you might try increasing the amount of pulmonary ionocytes to increase the amount of CFTR activity. The discovery could also make the task of trying to use gene therapy to correct the CFTR mutations a lot easier.
“I was surprised to spot potential paths to new therapies so quickly after doing the initial experiments,” says Lindsey Plasschaert, co-first author and postdoctoral researcher at NIBR.
This kind of research on the fundamental nature of different types of cells rarely yields insights with direct implications for drug discovery.
Technology opens new doors
Finding the new cell was no easy task. The team used a new technology called single-cell RNA sequencing to determine which genes were active in each individual cell in samples of airway tissue. Since gene activity defines a cell’s function (and thus its identity), they could use the technology to sort and catalog the many cells that allow us to make use of the air we breathe.
The secret – and the hard part – is being able to look at the gene activity of just one cell at a time. The NIBR scientists began working with the HMS scientists in 2015, about when the HMS scientists published a seminal paper on their single-cell RNA sequencing platform, which is called InDrops. Up to that point, researchers had been able to perform single-cell analysis of genetic activity on a few cells at a time, not the thousands of cells needed for a catalog like the one Jaffe envisioned for the airway.
InDrops enables scientists to capture individual cells in water droplets and then wrap them in oil so that the contents of the cells don’t mix. To look at only active genes, scientists must start with the cell’s messenger RNA. Messenger RNA delivers information encoded in the DNA to the machinery that produces proteins, and it is only made when a gene is active.
Ideally, scientists would sequence the messenger RNA to identify the source genes. But with no good method for sequencing RNA, it must be converted back into DNA. Doing this on many thousands of isolated cells would be a time-consuming and expensive task, so Klein’s lab figured out how to rapidly label the contents of the cell with tiny pieces of DNA that act as barcodes by isolating each cell in a tiny water droplet in oil.

“We are introducing the DNA into the droplets using squishy gels,” says Klein. “Each has about a billion little tags on it, which are then used to decorate all the material from the cell. Then we can break the droplets open, and the contents of each cell are now tagged with a different barcode. And we can read out the barcodes later and figure out which genetic material came from which cell.”
InDrops is now commercially available, but the Novartis researchers were able to access the technology for the airway project when it had just been invented. And Klein’s lab is on the cutting edge of analyzing the data that it produces. Graduate student Rapolas Zilionis, co-first author on the paper, handled this for the team along with Virginia Savova from the Klein lab.
“They analyzed the data and then fed it into an interactive viewer that allowed me to go through and look for differences in gene activity between the different cell types,” Plasschaert explains.
New cell pops out
Looking at that activity was essential to the catalog of cell types envisioned at the start of the project. The analysis revealed a couple clusters of cells that did not appear to be like anything in the scientific literature, which sent Plasschaert on a laborious journey to further characterize those cells.
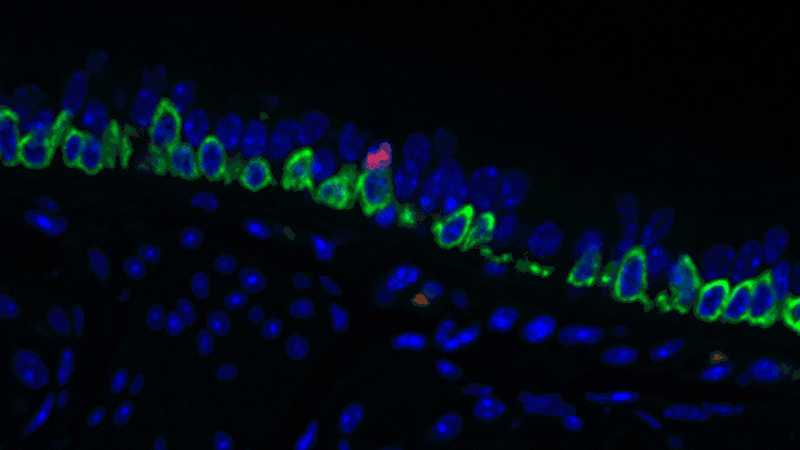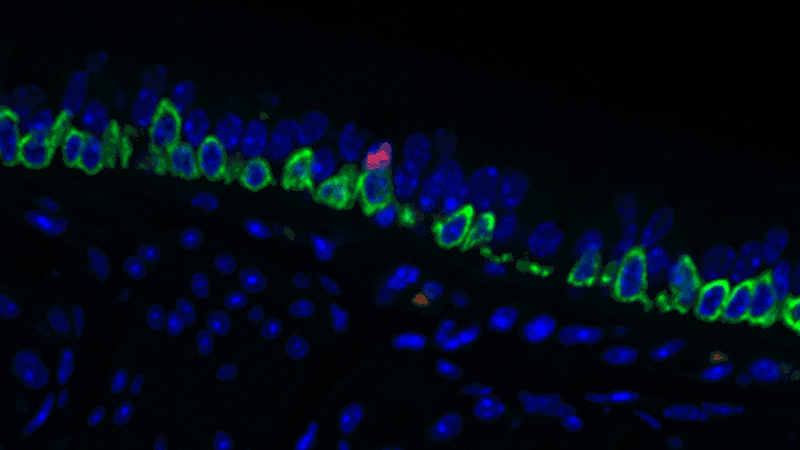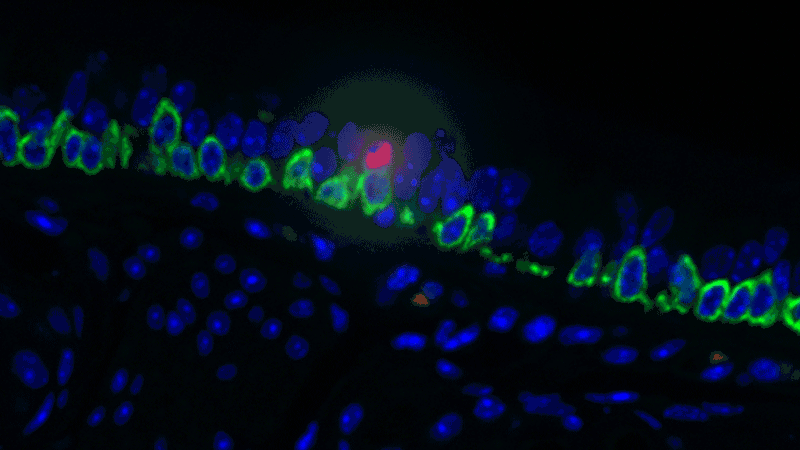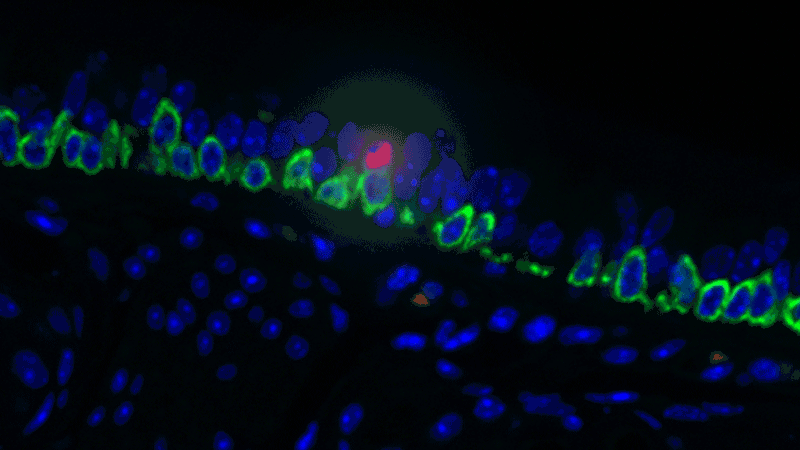
In a sense, it was like she was looking at a bowl of jelly beans, and she could see entirely new colors but had no idea what flavor they were (or, in the case of the cells, what function they performed). The experiments she designed showed that one set resembled the ionocytes – or cells that transport ions – found in some other tissues, most commonly in fish and frogs.
In fish, ionocytes transport ions to maintain an equilibrium with the water that surrounds them. In a bit of an evolutionary parallel, the cells discovered by the Novartis-Harvard team – pulmonary ionocytes – move ions at the interface between our tissues and the air around us. After considering various ways the ionocytes could do this, Plasschaert settled on CFTR activity.
The CFTR protein was known to transport ions. And Plasschaert conducted several experiments to show that the newly discovered pulmonary ionocytes have much more of the CFTR activity than another type of cell, ciliated cells, previously thought to be the main site of CFTR activity in the airway.
“The key finding turned out to be the high level of CFTR activity in the ionocytes and not in the ciliated cells people expected,” Plasschaert says. “This will help us target the functionally relevant cells for cystic fibrosis therapies going forward.”
Graduate student Zilionis says that from the beginning, he was excited about the project’s main goal of cataloging the types of cells that make up our airway. He was also surprised that it produced a result with therapeutic potential so quickly.
“We can now design ways to isolate pulmonary ionocytes,” he says. “We can get those cells in a dish and start doing things with them, looking for how they impact disease.”
Main image: Postdoctoral researcher Lindsey Plasschaert of Novartis and graduate student Rapolas Zilionis of Harvard Medical School review visual markers of a new airway cell type. Photo by PJ Kaszas
The co-first author on the paper, Rapolas Zilionis, is a visiting graduate student to the Klein lab from Vilnius University in Lithuania.
Learn how high-risk, high-reward science at Novartis yields #drugdiscovery targets for #cysticfibrosis.



